Targeting surface adhesins of pathogens has been receiving attention in recent years. Malaria, being a major killer, has been a target of intense investigation since last few decades. The identification of new drug targets and vaccine candidates to control the spread of this dreaded disease is an area of major thrust. In this context, the development of an algorithm dedicated to identify Plasmodial adhesins and antigens will serve to accelerate the ongoing research.
We have developed MAAP, a web server for the prediction of malarial adhesins and antigens. The MAAP algorithm uses 420 compositional properties and Support Vector Machines (SVM) to classify a given protein as either an adhesin or a non-adhesin.
MAAP accepts FASTA format protein sequences as input and provides an SVM based numeric score to these sequences, that is indicative of the probability of a malarial protein being an adhesin or an antigen.
Using MAAP, an efficiency of 0.86-0.89 has been achieved with sensitivity of 95.9% for Plasmodium falciparum and 100% for P. vivax and P. yoelii (when tested on known adhesin cases). The MAAP algorithm identifies most adhesins and antigens that are under intense investigations for vaccine development. In addition, several hypothetical proteins with potential adhesins and antigenic characteristics have been identified that can serve as new leads for further characterization.
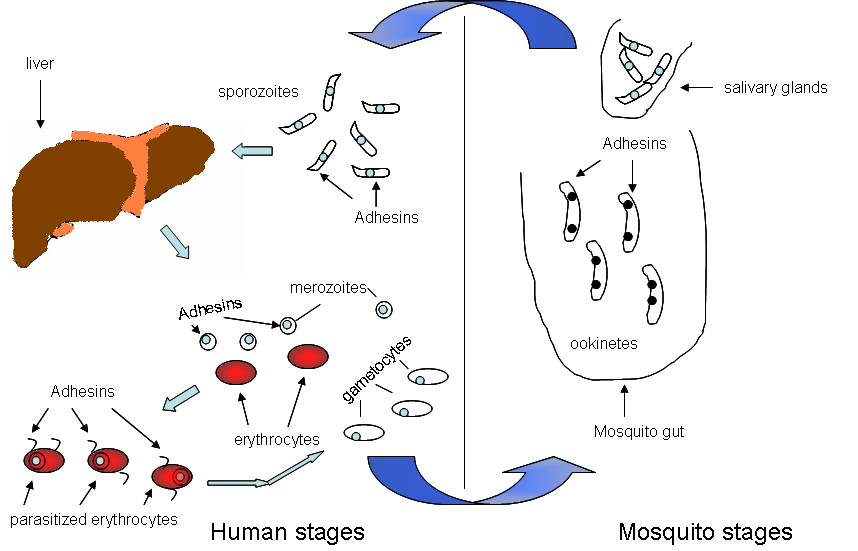 |
Figure. Adhesin surface proteins are required at various stages of the malaria parasite. The case for P. falciparum is shown. Since pathogens usually use a battery of adhesins for invasion, several of these adhesins await identification and characterization. Although the molecular details of interactions of adhesins at various stages likely differ from each other, in principle they could be identified using compositional properties that underlie the broad similarity characteristic of these proteins.
|
| 
